Projects
As the technology for gathering data becomes ever more powerful, the tools for analyzing the increasing volumes of information need to become more 'intelligent'. This data boom (particularly driven by the mapping of genomes) has led to the rapid development of machine learning algorithms.
The CLRC has developed a number of practically applicable algorithms (in partnership with both industrial and civil organisations) for a wide variety of problems. The versatility of machine learning algorithms allows them to be applied to a range of situations where ordinary statistical analytic methods cannot deal with the large volumes of data. Furthermore, the recent development of confidence machines means that predictions made are paired with a measure of confidence, allowing the human decision makers to better evaluate the analysis presented to them.
Below are a few examples of the application of CLRC algorithms. A list of current and past research grants can be found here.
Projects within the chemoinformatics domain
ExCAPE: Application of Uncertainty Quantification techniques to Compound Activity Prediction (virtual screening) for the Pharmaceutical industry.
Projects within the medical domain
For a collection of reports and publications from several these projects please visit the following page: CLRC reports on applications to proteomics and genomics.
Discovery of novel cancer serum biomarkers based on aberrant post-translational modifications of O-glycoproteins, O-PTM-Biomarkers, and their application to early detection of cancer
Cancer is a leading cause of death in developed countries, including Europe with one in three people being affected. Early detection accomplished through an efficient screening program remains the most promising approach to improve the long-term survival of cancer patients.
Therefore there is a pressing need for the development of biomarkers, which detect the early changes that lead to overt malignancy, particularly for cancers where clinical symptoms only appear when the cancer has progressed, and patients have poor survival (eg. ovarian, pancreatic, and lung cancer). This project addresses this problem.
Proteomic Analysis of the Human Serum Proteome for Population Screening, Diagnosis and Biomarker Discovery
In a joint project with University College London's Gynaecological Oncology and the Ludwig Institute for Cancer Research, the CLRC is helping to establish the potential for biomarker discovery and diagnosis of cancers and other disorders in large populations for high throughput protein analysis and pattern recognition.
Further information on this study can be found here.
Comparisons of Plasma Protocols
The team from the Proteomics Analysis of Human Serum Proteome study is collaborating with Ciphergen to analyse serum sample collection protocols. Reports from this study can be found below.
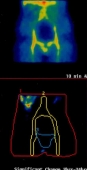
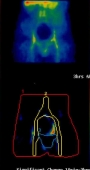
Ovarian Cancer - Adnexal mass categorising:
 |
 |
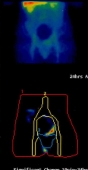 |
Working with St. Bartholomew's in London the CLRC used a similar system toperform a diagnostic of ovarian cancer, using a variety of symptoms in 287 patients to distinguish benign adnexal masses frommalignant.
The images to the right show a patients scan over a period of 24 hours, with the variation being analysed.
Abdominal pain diagnostic tool:
In partnership with Western General Hospital, Edinburgh, the CLRC developed a Bayesian algorithm for the diagnosis of abdominal pain in patients. The CLRC analysed data on 6,387 patients, each suffering abdominal pains described by one or more of 33 identified symptoms. Given this data, the learning machine (a G&T system) outputs a probability that the patient was suffering from each of 9 separate diseases.
As can be seen in the table on the right, in comparative testing the different versions of the G&T algorithms perform very favourably when compared to human predictions.
| Consultants | 76% |
| Registrars | 65% |
| Junior Doctors | 61% |
| G&T | 65% |
| G&T - simple bayes | 74% |
| G&T - CART | 64% |
Rate of successful diagnosis
of trained humans and learning algorithms. |
|
Veterinary Laboratories Agency project
A collaborative project with the Veterinary Laboratories Agency (VLA) entitled "Development and Application of Machine Learning Algorithms for the Analysis of Complex Veterinary Data Sets" has started in 2008.
Projects within other domains
LINK Hydrocarbon Reservoir Project:
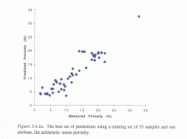
Working with Shell International, Cambridge Carbonates Ltd, British Gas Ltd and Petro-Canada, the CLRC assisted the Royal Holloway Department of Geology in the LINK Hydrocarbon Reservoirs Project. This project involved the creation of a permeability predictor database for use in oil exploration. The CLRC's contribution involved providing a learning machine to create a method of comparison, based on analysis of porosity and permeability data, for determining the porosity and permeability of unknown samples.
Again, use of computer learning methods scores over conventional programming and analysis in providing a self-correcting system instead of a flat database. It also frees the analysis from the restrictions of regular pattern-matching, allowing for a higher degree of 'guesswork' than is possible in conventional methods.
 |
 |
 |
|
Offender profiling:
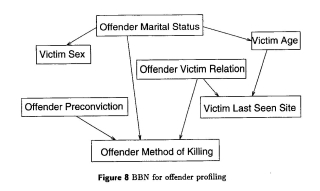
Using a database set up in 1986 by Derbyshire Constabulary and the input of detectives, the CLRC developed an offender profiling algorithm that allowed detectives to input a known variable (eg. Method of killing) to extrapolate further details (probabilities thereof) about the offender. Such a tool can help focus the limited resources of the investigating detectives. With recent incidents driving a call for improved police databases, such tools could become more prevalent in modern police work.
Tel/Fax : +44 (0)1784 443421 /439786

